Machine-Learned Collective Variables for Complex Biomolecular Systems
Leveraging machine learning to uncover low-dimensional, physically meaningful representations of rare molecular events such as protein folding and ligand binding.
Overview
Traditional molecular dynamics (MD) simulations of biomolecules are often constrained by the challenge of identifying an appropriate low-dimensional representation, or collective variable (CV), that effectively captures rare events such as protein folding, ligand binding, or conformational switching. Recent advances in machine learning (ML) provide powerful data-driven frameworks to automatically discover nonlinear CVs from simulation data, reducing the need for manual intuition and enabling deeper mechanistic understanding.
Key Concepts & Advantages
- » Automated Discovery: ML architectures such as autoencoders and variational autoencoders (VAEs) systematically compress high-dimensional atomic coordinates into a few latent variables that retain essential dynamical information.
- » Enhanced Sampling: These learned CVs can be combined with advanced sampling techniques — including Metadynamics, Umbrella Sampling, and Weighted Ensemble methods — to accelerate sampling of rare events and improve free energy estimations.
- » Physical Insight: ML-derived CVs reveal the most influential degrees of freedom, providing intuitive insight into transition mechanisms and structural heterogeneity in biomolecular systems.
Case Study: IceCoder — Machine Learning Framework for Ice Phase Classification
Identifying and classifying different phases of ice in molecular simulations is notoriously difficult due to
the subtle structural variations among crystalline and amorphous forms.
We developed IceCoder, a machine learning framework that integrates
Smooth Overlap of Atomic Positions (SOAP) descriptors with a
Variational Autoencoder (VAE) to learn a compact, two-dimensional latent representation of structural diversity in ice.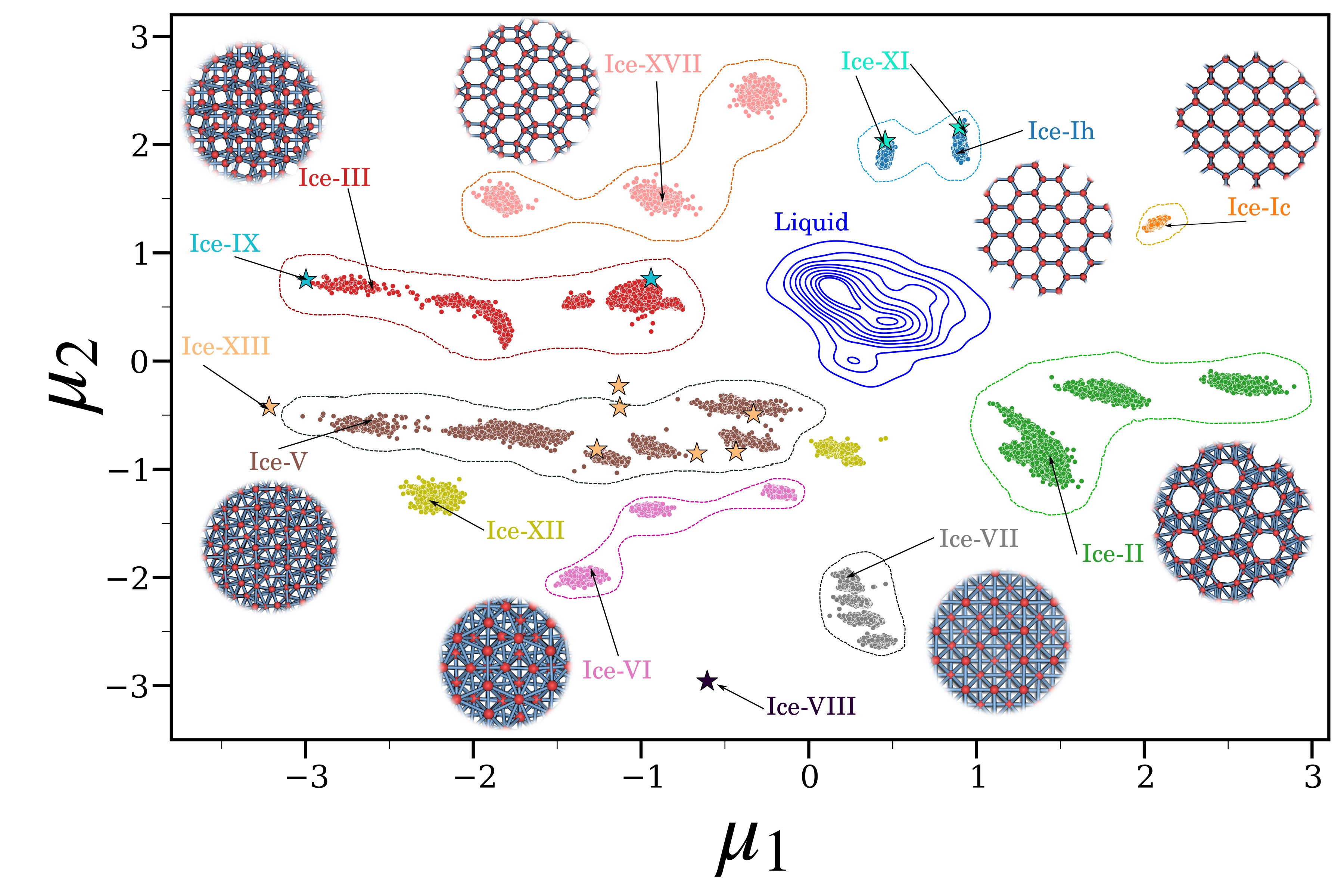
Trained on extensive molecular dynamics data, IceCoder successfully distinguishes between multiple crystalline
ice polymorphs and liquid water, even under thermal fluctuations. Beyond ice, this framework can be generalized
to identify polymorphs in other molecular crystals, offering a scalable and interpretable approach
to track nucleation, growth, and phase transitions with high fidelity and computational efficiency.
References:
Maity et al., Chem. Theory Comput. 2025, 21, 4, 1916–1928