Molecular dynamics (MD) simulations are a powerful tool for understanding molecular behavior, but simulating rare events—such as protein folding, ligand binding, or conformational transitions—remains computationally challenging due to long waiting times and high free-energy barriers. Recent advances in enhanced sampling methods have addressed these challenges by intelligently guiding simulations toward target states and efficiently estimating kinetic properties.
WeTICA: A Directed Search Weighted Ensemble Based Enhanced Sampling Method
WeTICA is a binless Weighted Ensemble (WE) algorithm designed to efficiently estimate rare event kinetics without the need for complex binning schemes.
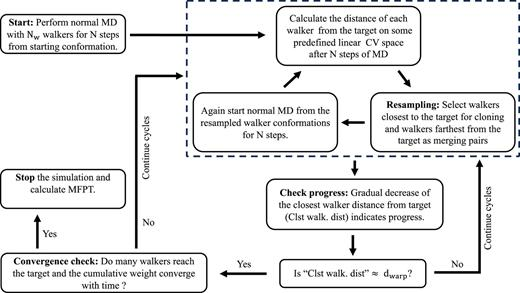
Diagram illustrating the WeTICA enhanced sampling method
Key Features
- » Uses low-dimensional Time-lagged Independent Component Analysis (TICA) projections as collective variables (CVs).
- » Enables the direct calculation of kinetic rates from biased trajectories.
- » Applied to unfolding of TC5b and TC10b Trp-cage mutants, as well as Protein G, with high accuracy.
- » Achieves convergence using an order of magnitude less simulation time than conventional WE approaches.
- » Can be extended to other linear or nonlinear CV spaces with improved walker selection criteria.
References
- Mitra et al., J. Chem. Phys. 162, 034106 (2025)
CoWERA: Coherence-based Weighted Ensemble Resampling Algorithm
Overview
CoWERA introduces temporal coherence-based resampling to prioritize trajectories that consistently make forward progress toward the target state.
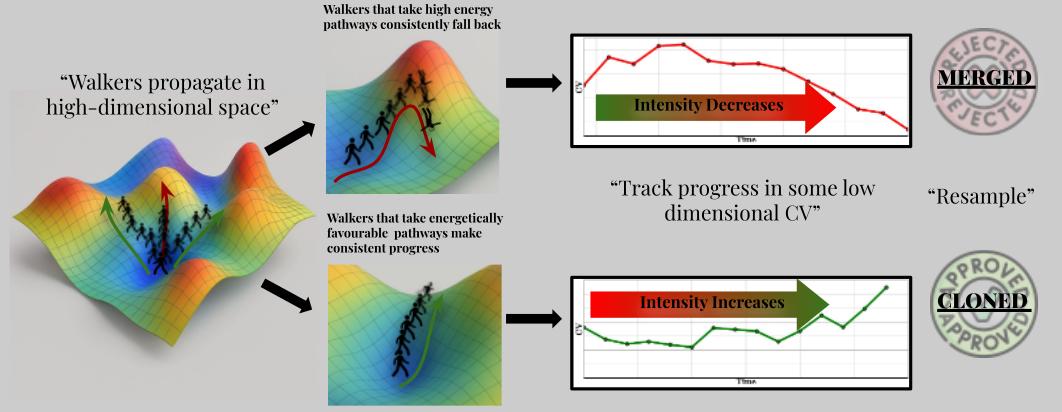
Key Features
- » Focuses computational effort on physically meaningful and productive pathways.
- » Reduces wasteful sampling in unproductive CV regions.
- » Provides faster convergence, lower variance, and more reliable kinetic estimates.
- » Avoids misleading guidance from absolute CV positions, ensuring kinetic relevance.
Applications
Protein folding, ligand unbinding, conformational switching, and other rare-event molecular processes where efficiency and kinetic accuracy are critical.
References
- Work in progress!
PathGennie: Rapid Generation of Rare Event Pathways
Overview
PathGennie uses a direction-guided adaptive sampling strategy with ultrashort monitored trajectories to quickly identify rare event pathways.
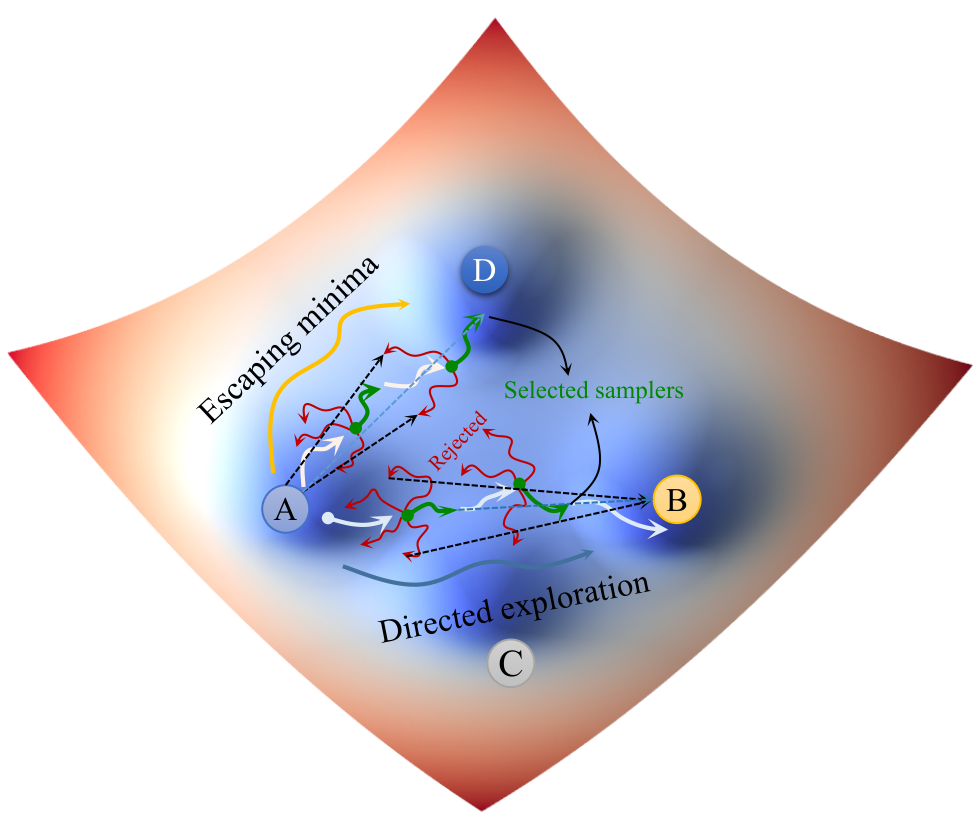
Key Features
- » Selectively propagates trajectories showing progress toward the target in a predefined CV space.
- » Handles ligand unbinding in T4 Lysozyme and Abl kinase, as well as protein folding (Trp-cage, Protein G).
- » Generates multiple competing pathways and estimates their relative populations.
- » Produces viable transition pathways in a few hundred picoseconds, suitable for further analysis using WE or TPS simulations.
References
- Work in progress!
Open-source code: PathGennie on GitHub